Arts & Photography
Audio CDs
Audiocassettes
Biographies & Memoirs
Business & Investing
Children's Books
Christianity
Comics & Graphic Novels
Computers & Internet
Cooking, Food & Wine
Entertainment
Gay & Lesbian
Health, Mind & Body
History
Home & Garden
Horror
Literature & Fiction
Mystery & Thrillers
Nonfiction
Outdoors & Nature
Parenting & Families
Professional & Technical
Reference
Religion & Spirituality
Romance
Science
Science Fiction & Fantasy
Sports
Teens
Travel
Women's Fiction
|
 |
Introduction to Computational Molecular Biology |
List Price: $98.95
Your Price: $98.95 |
 |
|
|
|
| Product Info |
Reviews |
<< 1 >>
Rating: 
Summary: Actually 3+
Review: Good for a beginner but lacks a lot of topics in bioinformatics
as well as a rigourous approach. But I must say the that the concepts in the book are made clear in a good style.
Rating: 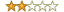
Summary: Not a good introduction book
Review: I do not think it is a good introduction book for biologists to learn computational biology. The authors should have used more figures and examples to illustrate the concepts. Also, I do not like the norrow margins of this book. I like to write comments and my understanding in the books when I read. There is simply no place to write.
Rating: 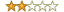
Summary: Not a good introduction book
Review: I do not think it is a good introduction book. The authors should have used more figures to illustrate the concepts. Also, I do not like the norrow margins of this book. I like to write comments and my understanding in the books. There is simply no place to write!
Rating: 
Summary: One of the really wonderful books in biocomputing.
Review: The book gives a brief introduction on the mathematical structures used, without getting lost in mathematical obscurities. The algorithmic representation of all models make it easy to implement the models. It is a book of great practical use. I very strongly recommend this book for biologists as well as computer scientists interested in the area of biocomputing.
Rating: 
Summary: Strong on algorithm analysis
Review: This is a good book on some of the algortihms used in bioinformatics. That term may not have been in wide use back in `97, leading to the title that suggests a lot more focus on the molecules than is in fact present.
The book starts with two brief chapters on biology and mathematical basics - graph theory and algorithm analysis. I'm sorry to say that these really are too brief. A reader who comes in with little knowledge of either topic will probably leave with about the same level of knowledge.
After that, the authors give coverage of the basics, as `97 writers saw them: approximate string matching, fragment assembly, mapping, trees, and a discussion of the reversals that occur in DNA over evolutionary time. Each topic is presented carefully, in a number of variations, and with formal analysis of the algorithmic complexity. That last won't do much for the biologists in the crowd, but gives programmers a good idea of how each technique will behave as the problems grow larger (and they always do). The presentation generally stick with the most popular algorithms, emphasizing detailed presentation over breadth of coverage. Multiple alignment, in particular, could have used a lot more pages. Also, topics like assembly and restriction digests aren't at the forefront of analysis any more. They're important, but good algorithms exist in widely available tools, and more advanced analyses tend to attract more attention these days. The section on genome rearrangements is quite good, but seems to stand alone - it could have been one input into tree building, but the authors don't draw any clear relationships between the reorderings and any other problems.
The section on structure prediction is definitely showing its age. RNA structure prediction has come a long way since this was written, and protein structure prediction has come even farther. Discussions of the basic ideas are good, but a more recent reader will want a lot more development. The final section, on using DNA as a material for performing computations, is interesting but hardly mainstream. The authors use one or two problems as case studies, but don't present the kinds of tools that can be applied to lots of different problems, just a few point solutions.
This book's value, today, lies mostly in the clarity of its complexity analysis and in the pseudocode that gives a programmer a step in the right direction. It has a few unusual items, like a highly generalized gap model for approximate string matching. On the whole, though, more recent books present most of the same material at least as well, and cover more up-to-date topics.
When it was new, I might have given this book a five star rating. Times have changed, though, and I have to rate this book among the others on the shelves now.
//wiredweird
Rating: 
Summary: Concise and to-the-point
Review: This is a really nice introduction to the most commonly used algorithms in bioinformatics. It is not really a general introduction to molecular biology or to computer science, and to make best use of this book a reader probably needs some prior exposure to both. But, for someone who has had a basic course in say genetics, and a basic programming course that covers simple data structures and algorithms etc., this volume provides all they will need to understand what is really happening when they run a BLAST search, for instance. A serious computer-science type person will probably not find the alogorithms described here very interesting, because they aren't meant to be elegant or interesting, just useful. I think a reader would have to have some direct interest in bioinformatics per se in order to enjoy this book. One thing that I find particularly nice is that the length of the chapters is just right so that you can read through a chapter in a single sitting, and because the chapters are largely independent of one another, its a handy book to have around and pick up when one has a little spare time. I recommend it very strongly.
Rating: 
Summary: Concise and to-the-point
Review: This is a really nice introduction to the most commonly used algorithms in bioinformatics. It is not really a general introduction to molecular biology or to computer science, and to make best use of this book a reader probably needs some prior exposure to both. But, for someone who has had a basic course in say genetics, and a basic programming course that covers simple data structures and algorithms etc., this volume provides all they will need to understand what is really happening when they run a BLAST search, for instance. A serious computer-science type person will probably not find the alogorithms described here very interesting, because they aren't meant to be elegant or interesting, just useful. I think a reader would have to have some direct interest in bioinformatics per se in order to enjoy this book. One thing that I find particularly nice is that the length of the chapters is just right so that you can read through a chapter in a single sitting, and because the chapters are largely independent of one another, its a handy book to have around and pick up when one has a little spare time. I recommend it very strongly.
<< 1 >>
|
|
|
|